Efrat Shema’s team from the Weizmann Institute of Science in Israel published a research paper titled “Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics” in the journal Nature Biotechnology (IF=68). This study developed a single-molecule multiparameter assay to comprehensively analyze epigenetics of plasma-isolated nucleosomes (EPINUC) information, including DNA methylation and cancer-specific protein biomarkers thing.
The tissue of origin of colorectal, pancreatic, lung and breast tumors was revealed by combining EPINUC with direct single-molecule DNA sequencing. In conclusion, EPINUC provides an effective means to obtain clinically relevant multi-layered information from limited (<1 ml) liquid biopsy material.
Analysis of cell-free DNA (cfDNA) in plasma provides information about pathological processes in the body. Blood cfDNA exists in the form of nucleosomes that maintain its tissue- and cancer-specific epigenetic state. In the plasma and serum of healthy individuals, circulating cfDNA originates primarily from the death of normal blood cells. However, in individuals with cancer, a portion of the cfDNA originates from the tumor, called circulating tumor DNA (ctDNA).
cfDNA in plasma occurs primarily in the form of nucleosomes (cfNucleosomes), the basic units of chromatin consisting of octamers surrounding a core histone protein and about 150 base pairs. Although abundant epigenetic information exists in plasma, this information has so far been mostly difficult to obtain. The main reason is that they require a large amount of input material and often only measure monolayer information (i.e. DNA methylation, individual histone modifications or nucleosome occupancy, etc.). Therefore, there is an urgent need for high-resolution methods that integrate information from multiple parameters across different types of analytes.
In this study, a single-molecule-based liquid biopsy method was developed to analyze multiple epigenetic parameters from <1 ml plasma samples, termed Epigenetics of Nucleosome Separation in Plasma (EPINUC). The technique is based on a single-molecule system that images combinatorial histone modifications by total internal reflection (TIRF) microscopy. TIRF provides a powerful method to detect single fluorescent molecules in the range of about 100 nm from the solid surface.
To capture nucleosomes from plasma, the researchers developed highly efficient enzymatic reactions to fluorescently labeled and polyadenylated nucleosomes. The tailed intact nucleosomes were then immobilized on PEGylated surfaces by hybridization, and their post-translational modification (PTM) status was recorded by TIRF imaging with fluorescently labeled antibodies. Using the narrow excitation range of TIRF to image antibody binding and dissociation from target PTMs within 90 minutes, this integration ensures maximum detection of histone modifications.
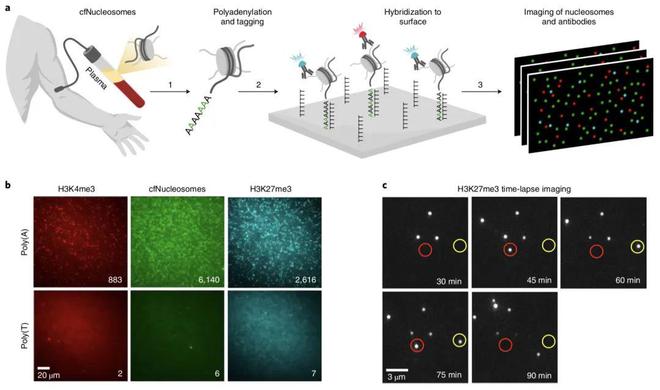
In addition, the system provides sensitive and quantitative data on plasma proteins, including detection of non-secreted tumor-specific proteins such as mutated p53. EPINUC analyzed a panel of 63 colorectal cancer, 10 pancreatic cancer, and 33 healthy plasma samples and detected cancer with high accuracy and sensitivity, even at an early stage.
Not only that, but the researchers combined the molecular biology of cancer with artificial intelligence algorithms to analyze these two large datasets using machine learning. This analysis is not only for all of these cancer markers, but also for their combination and the relationship between them. The results showed that the algorithm was 92 percent accurate when applied to studies that differentiated between healthy and patient populations.
Overall, this study developed a liquid biopsy method for comprehensive analysis of the epigenetics of plasma isolated nucleosomes (EPINUC) using <1 ml of blood, which allows logarithmic analysis of plasma isolated nucleosomes by single-molecule imaging. High-resolution detection of six active and repressive histone modifications and their ratios and combinatorial patterns on millions of single nucleosomes is not achievable with conventional techniques. In the future, EPINUC can be used as an effective and convenient detection method for diagnosing various cancers, or other diseases that leave traces in the blood, which will provide great help for clinical diagnosis of diseases.



GIPHY App Key not set. Please check settings